Software
This is a selection of open-source and research-related software I’ve developed. You can also find my GitHub for details.
Highlighted Open-Source Libraries
Bayex: Bayesian Optimization in JAX
PCAx: Differentiable PCA in JAX

t-SNEx: Minimal t-SNE in JAX

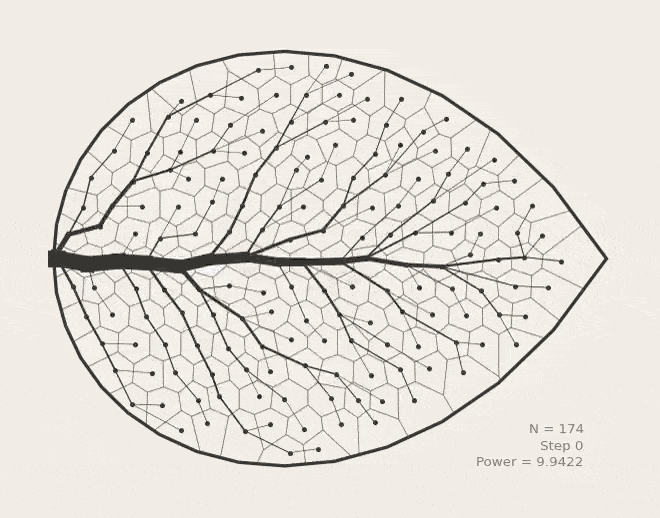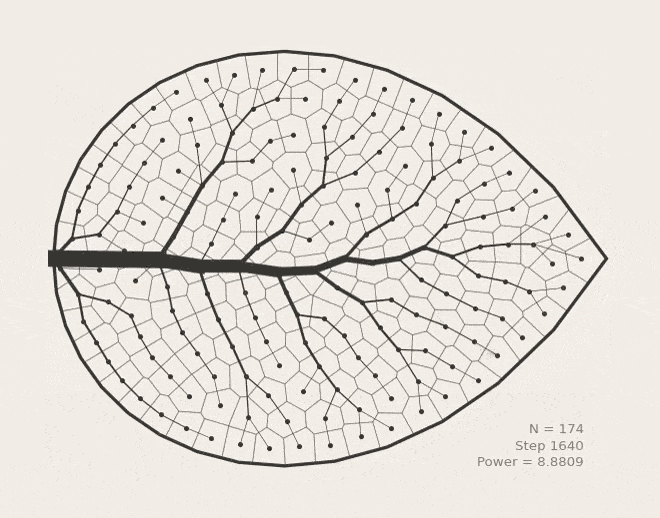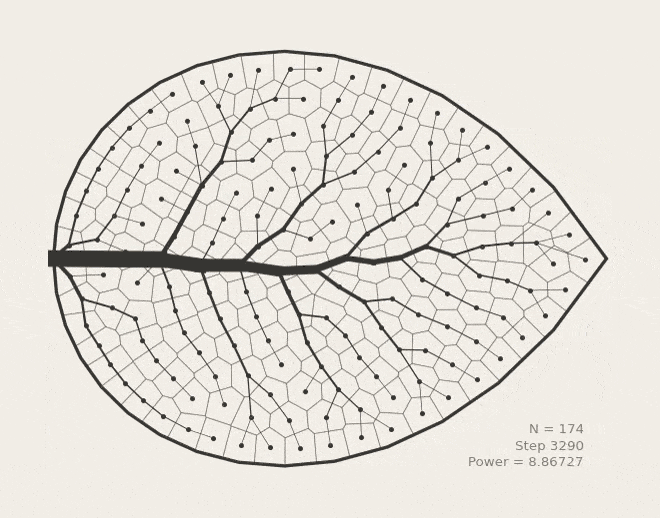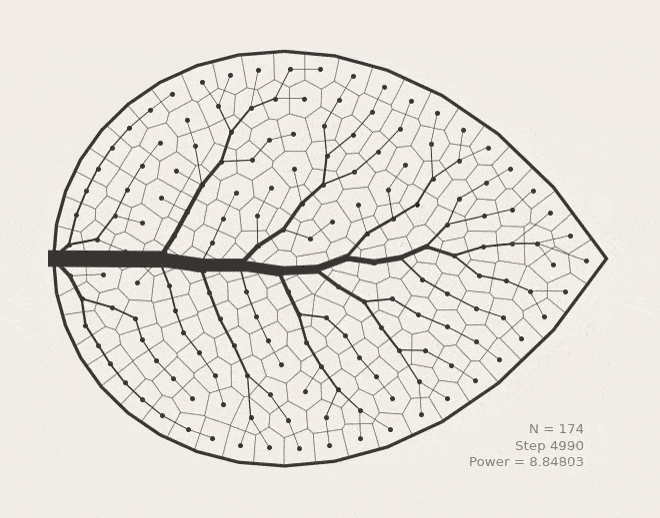
BoundVor: Bounded Voronoi Tessellation
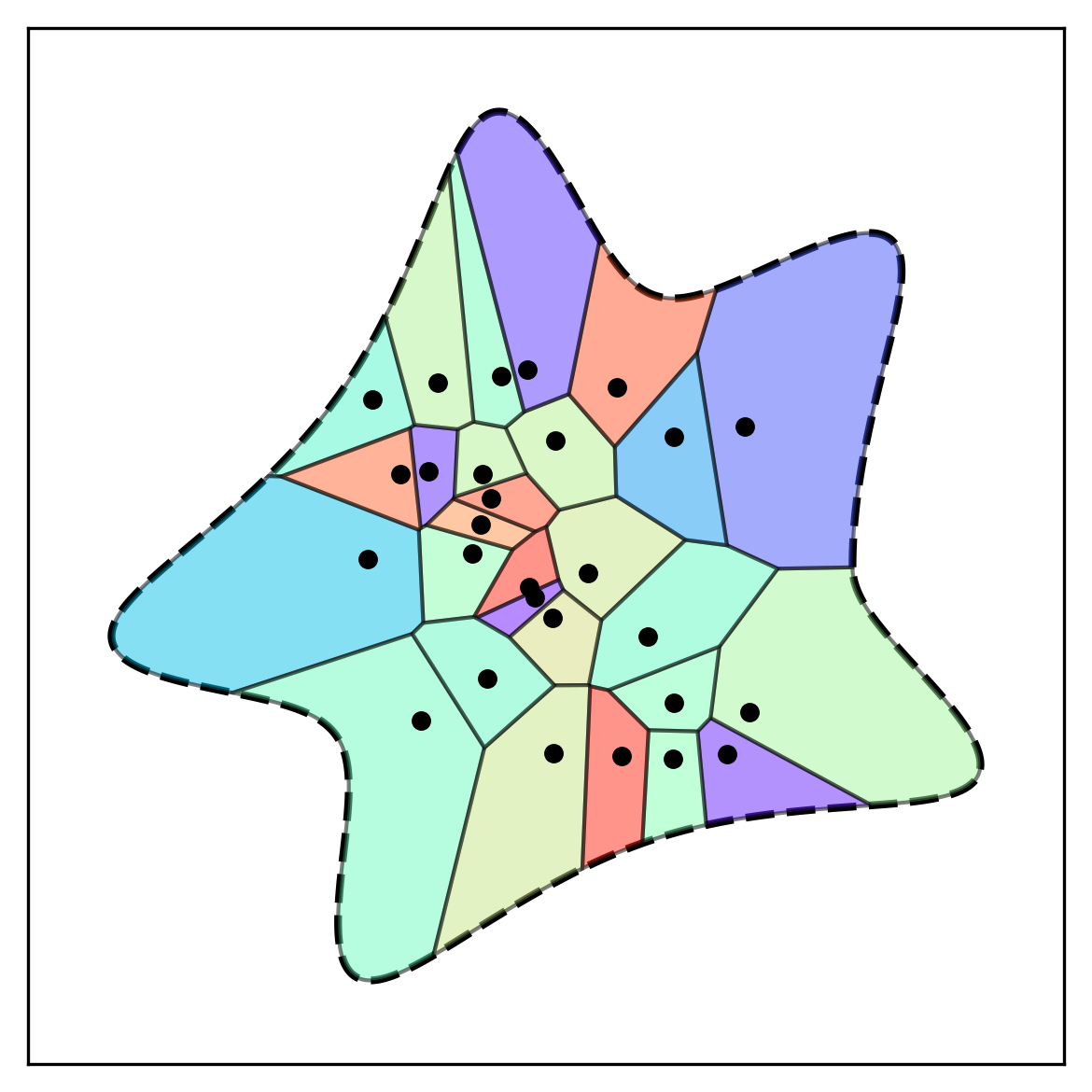
scipy.spatial.Voronoi to clip diagrams to arbitrary polygons. Useful for modeling bounded spatial domains in physical systems.
Notata: Scientific Logging

Highlighted Research Code
De(ep)tangle: High-Density Worm Tracking

ChemoXRL: Reinforcement Learning for Chemotaxis
Recloc: Receptor Clustering and Sensing Accuracy

GradNodes: Adaptive Node in Transport Networks